Lung Nodule Detection
Lung-nodule detection is the process of identifying and localizing nodules in lung CT scans for early diagnosis of lung cancer.
Papers and Code
Lung Nodule Image Synthesis Driven by Two-Stage Generative Adversarial Networks
Feb 02, 2026The limited sample size and insufficient diversity of lung nodule CT datasets severely restrict the performance and generalization ability of detection models. Existing methods generate images with insufficient diversity and controllability, suffering from issues such as monotonous texture features and distorted anatomical structures. Therefore, we propose a two-stage generative adversarial network (TSGAN) to enhance the diversity and spatial controllability of synthetic data by decoupling the morphological structure and texture features of lung nodules. In the first stage, StyleGAN is used to generate semantic segmentation mask images, encoding lung nodules and tissue backgrounds to control the anatomical structure of lung nodule images; The second stage uses the DL-Pix2Pix model to translate the mask map into CT images, employing local importance attention to capture local features, while utilizing dynamic weight multi-head window attention to enhance the modeling capability of lung nodule texture and background. Compared to the original dataset, the accuracy improved by 4.6% and mAP by 4% on the LUNA16 dataset. Experimental results demonstrate that TSGAN can enhance the quality of synthetic images and the performance of detection models.
Tri-Reader: An Open-Access, Multi-Stage AI Pipeline for First-Pass Lung Nodule Annotation in Screening CT
Jan 27, 2026Using multiple open-access models trained on public datasets, we developed Tri-Reader, a comprehensive, freely available pipeline that integrates lung segmentation, nodule detection, and malignancy classification into a unified tri-stage workflow. The pipeline is designed to prioritize sensitivity while reducing the candidate burden for annotators. To ensure accuracy and generalizability across diverse practices, we evaluated Tri-Reader on multiple internal and external datasets as compared with expert annotations and dataset-provided reference standards.
DGSAN: Dual-Graph Spatiotemporal Attention Network for Pulmonary Nodule Malignancy Prediction
Dec 24, 2025Lung cancer continues to be the leading cause of cancer-related deaths globally. Early detection and diagnosis of pulmonary nodules are essential for improving patient survival rates. Although previous research has integrated multimodal and multi-temporal information, outperforming single modality and single time point, the fusion methods are limited to inefficient vector concatenation and simple mutual attention, highlighting the need for more effective multimodal information fusion. To address these challenges, we introduce a Dual-Graph Spatiotemporal Attention Network, which leverages temporal variations and multimodal data to enhance the accuracy of predictions. Our methodology involves developing a Global-Local Feature Encoder to better capture the local, global, and fused characteristics of pulmonary nodules. Additionally, a Dual-Graph Construction method organizes multimodal features into inter-modal and intra-modal graphs. Furthermore, a Hierarchical Cross-Modal Graph Fusion Module is introduced to refine feature integration. We also compiled a novel multimodal dataset named the NLST-cmst dataset as a comprehensive source of support for related research. Our extensive experiments, conducted on both the NLST-cmst and curated CSTL-derived datasets, demonstrate that our DGSAN significantly outperforms state-of-the-art methods in classifying pulmonary nodules with exceptional computational efficiency.
Fairness Evaluation of Risk Estimation Models for Lung Cancer Screening
Dec 23, 2025Lung cancer is the leading cause of cancer-related mortality in adults worldwide. Screening high-risk individuals with annual low-dose CT (LDCT) can support earlier detection and reduce deaths, but widespread implementation may strain the already limited radiology workforce. AI models have shown potential in estimating lung cancer risk from LDCT scans. However, high-risk populations for lung cancer are diverse, and these models' performance across demographic groups remains an open question. In this study, we drew on the considerations on confounding factors and ethically significant biases outlined in the JustEFAB framework to evaluate potential performance disparities and fairness in two deep learning risk estimation models for lung cancer screening: the Sybil lung cancer risk model and the Venkadesh21 nodule risk estimator. We also examined disparities in the PanCan2b logistic regression model recommended in the British Thoracic Society nodule management guideline. Both deep learning models were trained on data from the US-based National Lung Screening Trial (NLST), and assessed on a held-out NLST validation set. We evaluated AUROC, sensitivity, and specificity across demographic subgroups, and explored potential confounding from clinical risk factors. We observed a statistically significant AUROC difference in Sybil's performance between women (0.88, 95% CI: 0.86, 0.90) and men (0.81, 95% CI: 0.78, 0.84, p < .001). At 90% specificity, Venkadesh21 showed lower sensitivity for Black (0.39, 95% CI: 0.23, 0.59) than White participants (0.69, 95% CI: 0.65, 0.73). These differences were not explained by available clinical confounders and thus may be classified as unfair biases according to JustEFAB. Our findings highlight the importance of improving and monitoring model performance across underrepresented subgroups, and further research on algorithmic fairness, in lung cancer screening.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2025:025
NodMAISI: Nodule-Oriented Medical AI for Synthetic Imaging
Dec 19, 2025Objective: Although medical imaging datasets are increasingly available, abnormal and annotation-intensive findings critical to lung cancer screening, particularly small pulmonary nodules, remain underrepresented and inconsistently curated. Methods: We introduce NodMAISI, an anatomically constrained, nodule-oriented CT synthesis and augmentation framework trained on a unified multi-source cohort (7,042 patients, 8,841 CTs, 14,444 nodules). The framework integrates: (i) a standardized curation and annotation pipeline linking each CT with organ masks and nodule-level annotations, (ii) a ControlNet-conditioned rectified-flow generator built on MAISI-v2's foundational blocks to enforce anatomy- and lesion-consistent synthesis, and (iii) lesion-aware augmentation that perturbs nodule masks (controlled shrinkage) while preserving surrounding anatomy to generate paired CT variants. Results: Across six public test datasets, NodMAISI improved distributional fidelity relative to MAISI-v2 (real-to-synthetic FID range 1.18 to 2.99 vs 1.69 to 5.21). In lesion detectability analysis using a MONAI nodule detector, NodMAISI substantially increased average sensitivity and more closely matched clinical scans (IMD-CT: 0.69 vs 0.39; DLCS24: 0.63 vs 0.20), with the largest gains for sub-centimeter nodules where MAISI-v2 frequently failed to reproduce the conditioned lesion. In downstream nodule-level malignancy classification trained on LUNA25 and externally evaluated on LUNA16, LNDbv4, and DLCS24, NodMAISI augmentation improved AUC by 0.07 to 0.11 at <=20% clinical data and by 0.12 to 0.21 at 10%, consistently narrowing the performance gap under data scarcity.
Scale-Aware Curriculum Learning for Ddata-Efficient Lung Nodule Detection with YOLOv11
Oct 30, 2025Lung nodule detection in chest CT is crucial for early lung cancer diagnosis, yet existing deep learning approaches face challenges when deployed in clinical settings with limited annotated data. While curriculum learning has shown promise in improving model training, traditional static curriculum strategies fail in data-scarce scenarios. We propose Scale Adaptive Curriculum Learning (SACL), a novel training strategy that dynamically adjusts curriculum design based on available data scale. SACL introduces three key mechanisms:(1) adaptive epoch scheduling, (2) hard sample injection, and (3) scale-aware optimization. We evaluate SACL on the LUNA25 dataset using YOLOv11 as the base detector. Experimental results demonstrate that while SACL achieves comparable performance to static curriculum learning on the full dataset in mAP50, it shows significant advantages under data-limited conditions with 4.6%, 3.5%, and 2.0% improvements over baseline at 10%, 20%, and 50% of training data respectively. By enabling robust training across varying data scales without architectural modifications, SACL provides a practical solution for healthcare institutions to develop effective lung nodule detection systems despite limited annotation resources.
Multi-Attention Stacked Ensemble for Lung Cancer Detection in CT Scans
Jul 27, 2025



In this work, we address the challenge of binary lung nodule classification (benign vs malignant) using CT images by proposing a multi-level attention stacked ensemble of deep neural networks. Three pretrained backbones - EfficientNet V2 S, MobileViT XXS, and DenseNet201 - are each adapted with a custom classification head tailored to 96 x 96 pixel inputs. A two-stage attention mechanism learns both model-wise and class-wise importance scores from concatenated logits, and a lightweight meta-learner refines the final prediction. To mitigate class imbalance and improve generalization, we employ dynamic focal loss with empirically calculated class weights, MixUp augmentation during training, and test-time augmentation at inference. Experiments on the LIDC-IDRI dataset demonstrate exceptional performance, achieving 98.09 accuracy and 0.9961 AUC, representing a 35 percent reduction in error rate compared to state-of-the-art methods. The model exhibits balanced performance across sensitivity (98.73) and specificity (98.96), with particularly strong results on challenging cases where radiologist disagreement was high. Statistical significance testing confirms the robustness of these improvements across multiple experimental runs. Our approach can serve as a robust, automated aid for radiologists in lung cancer screening.
Lung Nodule-SSM: Self-Supervised Lung Nodule Detection and Classification in Thoracic CT Images
May 21, 2025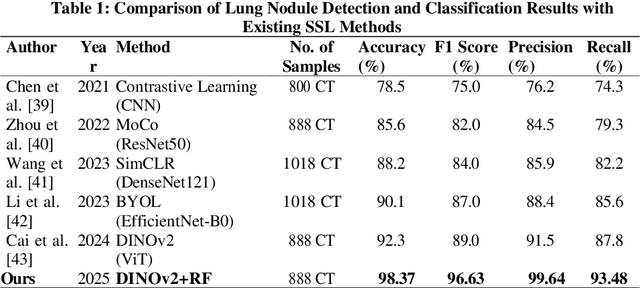
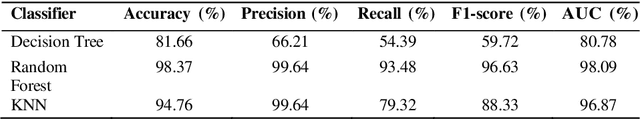
Lung cancer remains among the deadliest types of cancer in recent decades, and early lung nodule detection is crucial for improving patient outcomes. The limited availability of annotated medical imaging data remains a bottleneck in developing accurate computer-aided diagnosis (CAD) systems. Self-supervised learning can help leverage large amounts of unlabeled data to develop more robust CAD systems. With the recent advent of transformer-based architecture and their ability to generalize to unseen tasks, there has been an effort within the healthcare community to adapt them to various medical downstream tasks. Thus, we propose a novel "LungNodule-SSM" method, which utilizes selfsupervised learning with DINOv2 as a backbone to enhance lung nodule detection and classification without annotated data. Our methodology has two stages: firstly, the DINOv2 model is pre-trained on unlabeled CT scans to learn robust feature representations, then secondly, these features are fine-tuned using transformer-based architectures for lesionlevel detection and accurate lung nodule diagnosis. The proposed method has been evaluated on the challenging LUNA 16 dataset, consisting of 888 CT scans, and compared with SOTA methods. Our experimental results show the superiority of our proposed method with an accuracy of 98.37%, explaining its effectiveness in lung nodule detection. The source code, datasets, and pre-processed data can be accessed using the link:https://github.com/EMeRALDsNRPU/Lung-Nodule-SSM-Self-Supervised-Lung-Nodule-Detection-and-Classification/tree/main
An Efficient Approach to Detecting Lung Nodules Using Swin Transformer
Mar 03, 2025Lung cancer has the highest rate of cancer-caused deaths, and early-stage diagnosis could increase the survival rate. Lung nodules are common indicators of lung cancer, making their detection crucial. Various lung nodule detection models exist, but many lack efficiency. Hence, we propose a more efficient approach by leveraging 2D CT slices, reducing computational load and complexity in training and inference. We employ the tiny version of Swin Transformer to benefit from Vision Transformers (ViT) while maintaining low computational cost. A Feature Pyramid Network is added to enhance detection, particularly for small nodules. Additionally, Transfer Learning is used to accelerate training. Our experimental results show that the proposed model outperforms state-of-the-art methods, achieving higher mAP and mAR for small nodules by 1.3% and 1.6%, respectively. Overall, our model achieves the highest mAP of 94.7% and mAR of 94.9%.
A Narrative Review on Large AI Models in Lung Cancer Screening, Diagnosis, and Treatment Planning
Jun 08, 2025Lung cancer remains one of the most prevalent and fatal diseases worldwide, demanding accurate and timely diagnosis and treatment. Recent advancements in large AI models have significantly enhanced medical image understanding and clinical decision-making. This review systematically surveys the state-of-the-art in applying large AI models to lung cancer screening, diagnosis, prognosis, and treatment. We categorize existing models into modality-specific encoders, encoder-decoder frameworks, and joint encoder architectures, highlighting key examples such as CLIP, BLIP, Flamingo, BioViL-T, and GLoRIA. We further examine their performance in multimodal learning tasks using benchmark datasets like LIDC-IDRI, NLST, and MIMIC-CXR. Applications span pulmonary nodule detection, gene mutation prediction, multi-omics integration, and personalized treatment planning, with emerging evidence of clinical deployment and validation. Finally, we discuss current limitations in generalizability, interpretability, and regulatory compliance, proposing future directions for building scalable, explainable, and clinically integrated AI systems. Our review underscores the transformative potential of large AI models to personalize and optimize lung cancer care.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge